Sulfur Metabolism
Plants assimilate sulfate and synthesize a number of S-containing bioactive metabolites,
such as cysteine, methionine, glutathione, vitamins, and sulfolipids. In addition
to these essential metabolites, sulfated polysaccharides, sulfated peptides/proteins
and S-containing phytoalexins play critical roles in organogenesis, pathogenesis and
stress mitigation. Sulfate transporters and sulfate assimilatory enzymes are spatiotemporally
expressed and controlled in a demand-driven manner to efficiently utilize sulfate
for biosynthesis of S-containing metabolites. Because of the biological importance
of S in nature, our research is aimed at elucidating the molecular mechanisms of sulfate
transport, S metabolism, and S sensing and signaling in plants.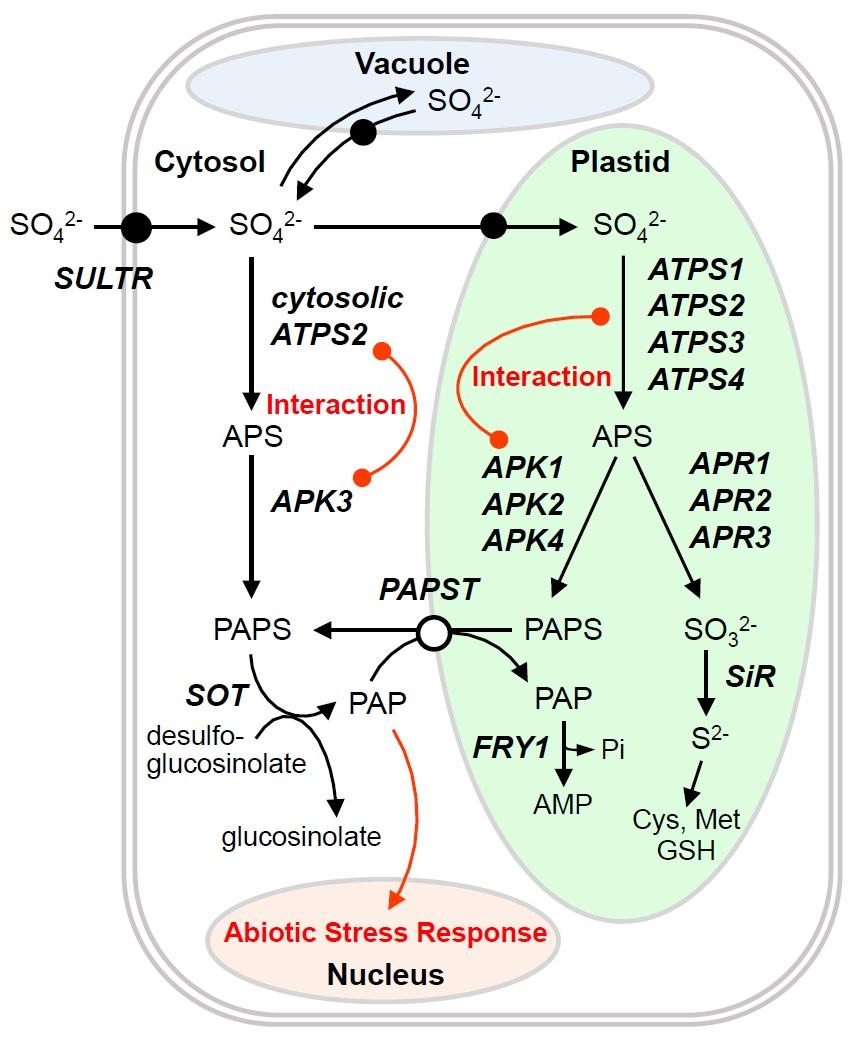
Our current NSF-funded project is focused on S-sensing and signaling mechanisms hypothesized to be associated with metabolic flux partitioning of the sulfate assimilation pathways in plastids and cytosol. This research project aims to identify new S-sensing functions of ATP sulfurylase (ATPS) and adenosine 5’-phosphosulfate kinase (APK), the first two pathway enzymes for the biosynthesis of adenosine 5’-phosphosulfate (APS) and 3’-phosphoadenosine 5’-phosphosulfate (PAPS). Our recent finding of plastid-cytosol dual localization of ATPS2 in Arabidopsis resolved a long-standing unanswered question on subcellular pathway compartmentalization of sulfate assimilation. Our preliminary studies using transgenic lines overexpressing the plastid-localizing ATPS1 further indicate that reinforcement of the plastid APS biosynthetic pathway causes a transcriptome-wide repression of S-responsive genes in Arabidopsis. Furthermore, the assembly of ATPS-APK protein complexes is suggested to be pivotal for controlling APS/PAPS biosynthesis in plastids and cytosol. These initial observations indicate that organellar-specific expressions of ATPS and APK isoforms and formation of ATPS-APK protein complexes are key elements for flux partitioning of APS/PAPS biosynthesis and for S-sensing and signaling mechanisms. Furthermore, the fluxes of APS/PAPS biosynthesis influence abiotic stress signaling mechanisms, since PAPS is used as a sulfate donor in sulfation and generates 3’-phosphoadenosine 5’-phosphate (PAP), which is known as a signaling molecule involved in induction of drought and high-light stress responses.
This research project provides a unique perspective beyond S-metabolic pathway compartmentalization. The mechanistic principle of S-sensing, which is based on metabolic flux partitioning of APS/PAPS biosynthesis and ATPS-APK complex formation, opens a new avenue towards understanding of nutrient sensing and stress signaling mechanisms.