Small Signaling Peptide Pathways

Plants modify root system architecture in response to give efficient access to nutrient resources that are unevenly distributed in the soil environment. Among the macronutrients required for plant growth, N has profound effects on developmental mechanisms. Lateral roots proliferate in local N-rich soil patches, but are restricted under prolonged N-deficiency, suggesting that prevention mechanisms act to economize the cost for root development and avoid the risks of extending the root systems into N-poor environments. To gain further insights into nutrient-responsive mechanisms affecting plant root system architecture, the Takahashi lab works on research projects that are aimed at identifying small signaling peptides involved in regulation of root and nodule development in Arabidopsis and Medicago species.
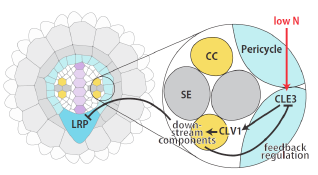
Our group recently discovered that lateral root development is modulated by CLE small signaling peptides (CLE1, 3, 4 and 7) expressed in the pericycle under N deficiency, and by CLV1, the CLE peptide receptor, in the phloem companion cells in roots. CLE-CLV1 peptide-receptor modules are also known to regulate nodule development in legumes. In addition, parasitic nematodes are known to produce small peptides homologous to CLE for establishment of feeding sites in plant roots. These initial findings depict the importance of studying small signaling peptides controlling plant root and nodule development in response to N and other macronutrients. However, hundreds of small signaling peptides in the plant genome remain uncharacterized, and there is a critical need to implement a functional genomics-empowered research to explore their diverse biological functions.
The current NSF-funded project, which has been initiated in collaboration with the Noble Foundation (Ardmore, OK), aims to identify macronutrient-responsive small signaling peptides involved in regulation of root and nodule development in the legume Medicago truncatula. Small signaling peptides altering root system architecture and nodule formation in response to N, S, P and K availability will be identified through transcriptome analysis and phenotypic screening of chemically synthesized peptides or overexpression of peptide-coding genes. We are also taking advantage of collections of genetic mutants defective for genes encoding leucine-rich repeat receptor-like kinases (LRR-RLK) to identify mutants for small peptide receptors. The downstream networks of signaling events associated with the functions of peptide-receptor modules will be investigated by analyzing the transcriptome profiles of wild-type plants and peptide receptor mutants following peptide treatments or overexpression of peptide-coding genes.
This research project on small signaling peptides is expected to provide key information on regulatory components and molecular networks that are relevant to the morphological strategies of plant roots applied for nutrient acquisition. The outcome of this research will have a strong impact on formulating genetic engineering strategies for modifying macronutrient and water uptake capacities of plant roots.